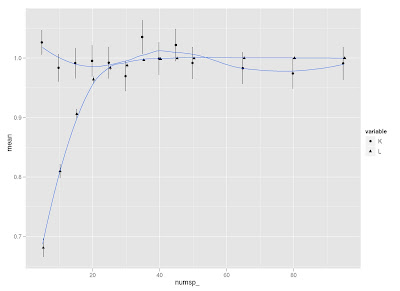
I did a little simulation to examine how K and lambda vary in response to tree size (and how they compare to each other on the same simulated trees). I use Liam Revell's functions fastBM to generate traits, and phylosig to measure phylogenetic signal.
Two observations:
First, it seems that lambda is more sensitive than K to tree size, but then lambda levels out at about 40 species, whereas K continues to vary around a mean of 1.
Second, K is more variable than lambda at all levels of tree size (compare standard error bars).
Does this make sense to those smart folks out there?
Recology HAS MOVED TO http://recology.info/. To get to the same blog post on the new site replace the http://r-ecology.blogspot.ca/ with http://recology.info/, but with the same ending, e,g. /2011/12/weecology-can-has-new-mammal-dataset.html (except remove the .html at the end)
Wednesday, May 18, 2011
Tuesday, May 17, 2011
A simple function for plotting phylogenies in ggplot2
UPDATE: Greg jordan has a much more elegant way of plotting trees with ggplot2. See his links in the comments below.
I wrote a simple function for plotting a phylogeny in ggplot2. However, it only handles a 3 species tree right now, as I haven't figured out how to generalize the approach to N species.
Any ideas on how to improve this?
I wrote a simple function for plotting a phylogeny in ggplot2. However, it only handles a 3 species tree right now, as I haven't figured out how to generalize the approach to N species.
Any ideas on how to improve this?
Friday, May 13, 2011
plyr's idata.frame VS. data.frame
I had seen the function idata.frame in plyr before, but not really tested it. From the plyr documentation:
"An immutable data frame works like an ordinary data frame, except that when you subset it, it
returns a reference to the original data frame, not a a copy. This makes subsetting substantially
faster and has a big impact when you are working with large datasets with many groups."
For example, although baseball is a data.frame, its immutable counterpart is a reference to it:
Here are a few comparisons of operations on normal data frames and immutable data frames. Immutable data frames don't work with the doBy package, but do work with aggregate in base functions. Overall, the speed gains using idata.frame are quite impressive - I will use it more often for sure.
Get the github code below here.
Here's the comparisons of idata.frames and data.frames:
Created by Pretty R at inside-R.org
"An immutable data frame works like an ordinary data frame, except that when you subset it, it
returns a reference to the original data frame, not a a copy. This makes subsetting substantially
faster and has a big impact when you are working with large datasets with many groups."
For example, although baseball is a data.frame, its immutable counterpart is a reference to it:
> idata.frame(baseball) <environment: 0x1022c74e8> attr(,"class") [1] "idf" "environment"
Here are a few comparisons of operations on normal data frames and immutable data frames. Immutable data frames don't work with the doBy package, but do work with aggregate in base functions. Overall, the speed gains using idata.frame are quite impressive - I will use it more often for sure.
Get the github code below here.
Here's the comparisons of idata.frames and data.frames:
> # load packages require(plyr); require(reshape2) > # Make immutable data frame baseball_i <- idata.frame(baseball) > # Example 1 - idata.frame more than twice as fast system.time( replicate(50, ddply( baseball, "year", summarise, mean(rbi))) ) user system elapsed 14.812 0.252 15.065 > system.time( replicate(50, ddply( baseball_i, "year", summarise, mean(rbi))) ) user system elapsed 6.895 0.020 6.915
> # Example 2 - Bummer, this does not work with idata.frame's> colwise(max, is.numeric) ( baseball ) # works
year stint g ab r h X2b X3b hr rbi sb cs bb so ibb hbp sh sf gidp
1 2007 4 165 705 177 257 64 28 73 NA NA NA 232 NA NA NA NA NA NA
> colwise(max, is.numeric) ( baseball_i ) # doesn't work
Error: is.data.frame(df) is not TRUE
> # Example 3 - idata.frame twice as fast system.time( replicate(100, baseball[baseball$year == "1884", ] ) ) user system elapsed 1.155 0.048 1.203 > system.time( replicate(100, baseball_i[baseball_i$year == "1884", ] ) ) user system elapsed 0.598 0.011 0.609
> # Example 4 - idata.frame faster system.time( replicate(50, melt(baseball[, 1:4], id = 1) ) ) user system elapsed 16.587 1.169 17.755 > system.time( replicate(50, melt(baseball_i[, 1:4], id = 1) ) ) user system elapsed 0.869 0.196 1.065
> # And you can go back to a data frame by d <- as.data.frame(baseball_i) str(d) 'data.frame': 21699 obs. of 23 variables: $ id : chr "ansonca01" "forceda01" "mathebo01" "startjo01" ... $ year : int 1871 1871 1871 1871 1871 1871 1871 1872 1872 1872 ... $ stint: int 1 1 1 1 1 1 1 1 1 1 ... $ team : chr "RC1" "WS3" "FW1" "NY2" ... $ lg : chr "" "" "" "" ... $ g : int 25 32 19 33 29 29 29 46 37 25 ... $ ab : int 120 162 89 161 128 146 145 217 174 130 ... $ r : int 29 45 15 35 35 40 36 60 26 40 ... $ h : int 39 45 24 58 45 47 37 90 46 53 ... $ X2b : int 11 9 3 5 3 6 5 10 3 11 ... $ X3b : int 3 4 1 1 7 5 7 7 0 0 ... $ hr : int 0 0 0 1 3 1 2 0 0 0 ... $ rbi : int 16 29 10 34 23 21 23 50 15 16 ... $ sb : int 6 8 2 4 3 2 2 6 0 2 ... $ cs : int 2 0 1 2 1 2 2 6 1 2 ... $ bb : int 2 4 2 3 1 4 9 16 1 1 ... $ so : int 1 0 0 0 0 1 1 3 1 0 ... $ ibb : int NA NA NA NA NA NA NA NA NA NA ... $ hbp : int NA NA NA NA NA NA NA NA NA NA ... $ sh : int NA NA NA NA NA NA NA NA NA NA ... $ sf : int NA NA NA NA NA NA NA NA NA NA ... $ gidp : int NA NA NA NA NA NA NA NA NA NA ... $ teamf: Factor w/ 132 levels "ALT","ANA","ARI",..: 99 127 51 79 35 35 122 86 16 122 ...
> # idata.frame doesn't work with the doBy package require(doBy) summaryBy(rbi ~ year, baseball_i, FUN=c(mean), na.rm=T) Error in as.vector(x, mode) : cannot coerce type 'environment' to vector of type 'any'
> # But idata.frame works with aggregate in base (but with minimal speed gains) # and aggregate is faster than ddply of course system.time( replicate(100, aggregate(rbi ~ year, baseball, mean) ) ) user system elapsed 4.117 0.423 4.541 > system.time( replicate(100, aggregate(rbi ~ year, baseball_i, mean) ) ) user system elapsed 3.908 0.383 4.291 > system.time( replicate(100, ddply( baseball_i, "year", summarise, mean(rbi)) ) ) user system elapsed 14.015 0.048 14.082
Thursday, May 12, 2011
google reader
I just realized that the gists code blocks don't show up in Google Reader, so you have to click the link to my blog to see the gists. Apologies for that!
-S
-S
Wednesday, May 11, 2011
Comparison of functions for comparative phylogenetics
With all the packages (and beta stage groups of functions) for comparative phylogenetics in R (tested here: picante, geiger, ape, motmot, Liam Revell's functions), I was simply interested in which functions to use in cases where multiple functions exist to do the same thing. I only show default settings, so perhaps these functions would differ under different parameter settings. [I am using a Mac 2.4 GHz i5, 4GB RAM]
Get motmot here: https://r-forge.r-project.org/R/?group_id=782
Get Liam Revell's functions here: http://anolis.oeb.harvard.edu/~liam/R-phylogenetics/
Created by Pretty R at inside-R.org
__________
It's hard to pick an overall winner because not all functions are available in all packages, but there are definitely some functions that are faster than others.
Get motmot here: https://r-forge.r-project.org/R/?group_id=782
Get Liam Revell's functions here: http://anolis.oeb.harvard.edu/~liam/R-phylogenetics/
> # Load require(motmot); require(geiger); require(picante) source("http://anolis.oeb.harvard.edu/~liam/R-phylogenetics/phylosig/v0.3/phylosig.R") source("http://anolis.oeb.harvard.edu/~liam/R-phylogenetics/fastBM/v0.4/fastBM.R") # Make tree tree <- rcoal(10)
# Transform branch lengths > system.time( replicate(1000, transformPhylo(tree, model = "lambda", lambda = 0.5)) ) # motmot user system elapsed 1.757 0.004 1.762 > system.time( replicate(1000, lambdaTree(tree, 0.9)) ) # geiger user system elapsed 3.708 0.008 3.716 > # motmot wins!!!
# Simulate trait evolutionsystem.time( replicate(1000, transformPhylo.sim(tree, model = "bm")) ) # motmot user system elapsed 3.732 0.007 3.741 > system.time( replicate(1000, rTraitCont(tree, model = "BM")) ) # ape user system elapsed 0.312 0.009 0.321 > system.time( replicate(1000, fastBM(tree)) ) # Revell user system elapsed 1.315 0.005 1.320 > # ape wins!!!
# Phylogenetically independent contrasts trait <- rnorm(10) names(trait) <- tree$tip.label > system.time( replicate(10000, pic.motmot(trait, tree)$contr[,1]) ) # motmot user system elapsed 3.062 0.007 3.070 > system.time( replicate(10000, pic(trait, tree)) ) # ape user system elapsed 2.846 0.007 2.853 > # ape wins!!!
# Phylogenetic signal, Blomberg's K > system.time( replicate(100, Kcalc(trait, tree)) ) # picante user system elapsed 1.311 0.005 1.316 > system.time( replicate(100, phylosig(tree, trait, method = "K")) ) # Revell user system elapsed 0.201 0.000 0.202 > # Liam Revell wins!!!
# Ancestral character state estimation > system.time( replicate(100, ace(trait, tree)$ace) ) # ape user system elapsed 4.988 0.018 5.007 > system.time( replicate(100, getAncStates(trait, tree)) ) # geiger user system elapsed 2.253 0.005 2.258 > # geiger wins!!!
__________
It's hard to pick an overall winner because not all functions are available in all packages, but there are definitely some functions that are faster than others.
Wednesday, May 4, 2011
RHIPE package in R for interfacing between Hadoop and R
RHIPE: An Interface Between Hadoop and R
Presented by Saptarshi Guha
Presented by Saptarshi Guha
And this review of methods for interfacing with Hadoop suggests R's RHIPE is quite nice.
Tuesday, May 3, 2011
Treebase trees from R
UPDATE: See Carl Boettiger's functions/package at Github for searching Treebase here.
Treebase is a great resource for phylogenetic trees, and has a nice interface for searching for certain types of trees. However, if you want to simply download a lot of trees for analyses (like that in Davies et al.), then you want to be able to access trees in bulk (I believe Treebase folks are working on an API though). I wrote some simple code for extracting trees from Treebase.org.
It reads an xml file of (in this case consensus) URL's for each tree, parses the xml, makes a vector of URL's, reads the nexus files with error checking, remove trees that gave errors, then a simple plot looking at metrics of the trees.
Is there an easier way to do this?
Treebase is a great resource for phylogenetic trees, and has a nice interface for searching for certain types of trees. However, if you want to simply download a lot of trees for analyses (like that in Davies et al.), then you want to be able to access trees in bulk (I believe Treebase folks are working on an API though). I wrote some simple code for extracting trees from Treebase.org.
It reads an xml file of (in this case consensus) URL's for each tree, parses the xml, makes a vector of URL's, reads the nexus files with error checking, remove trees that gave errors, then a simple plot looking at metrics of the trees.
Is there an easier way to do this?
Subscribe to:
Comments (Atom)