A new early online paper in
American Journal of Botany by Risa Sargent and colleagues suggests that plants are less sex deprived (pollen limited) in vernal pools that have more closely related plant species.
Vernal pools are (at least in my experience) small (to quite large) depressions that fill up with water with winter rains, and dry out completely in the summer. Vernal pool adapted plants flower in rings down the pool as the water dries up. Aquatic invertebrates and some herps can last through the summer by burrowing in the soil.
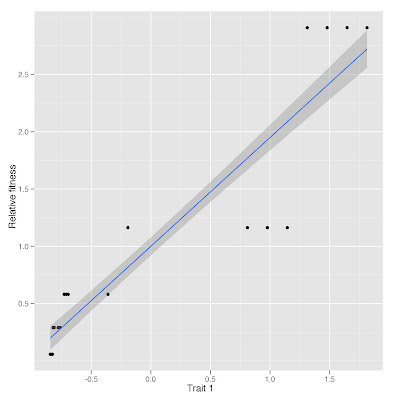
The study did hand pollination experiments with a focal species,
Lasthenia fremontii. They examined the relationship between these pollen limitation experiments and the relatedness of
L. fremontii to the rest of the plant community in each pool.
Plant species richness was not related to pollen limitation. Thus, at least in their study with vernal pools in California, relatedness to your plant neighbors has a greater impact than plant richness.
The great thing about vernal pools is that they are truly terrestrial islands of habitat, surrounded by inhospitable habitat for pool species. Many vernal pools are created artificially for habitat conservation easements (e.g.,
here). Perhaps someone can experimentally manipulate phylogenetic diversity in artificially created pools to really get at the causal links.
p.s. Ok, this post is not terribly R related, except for that this paper used R for some of their statistics.