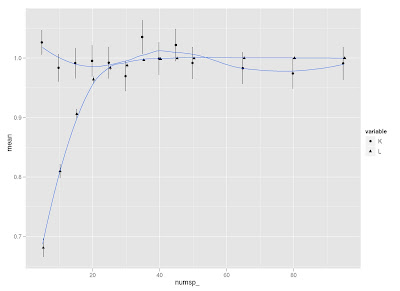
I did a little simulation to examine how K and lambda vary in response to tree size (and how they compare to each other on the same simulated trees). I use Liam Revell's functions fastBM to generate traits, and phylosig to measure phylogenetic signal.
Two observations:
First, it seems that lambda is more sensitive than K to tree size, but then lambda levels out at about 40 species, whereas K continues to vary around a mean of 1.
Second, K is more variable than lambda at all levels of tree size (compare standard error bars).
Does this make sense to those smart folks out there?
This comment has been removed by the author.
ReplyDelete